About
HomolWat (HW) is a free tool to incorporate internal water molecules to GPCR structures based on homologous structures with determined water molecules.
Usage
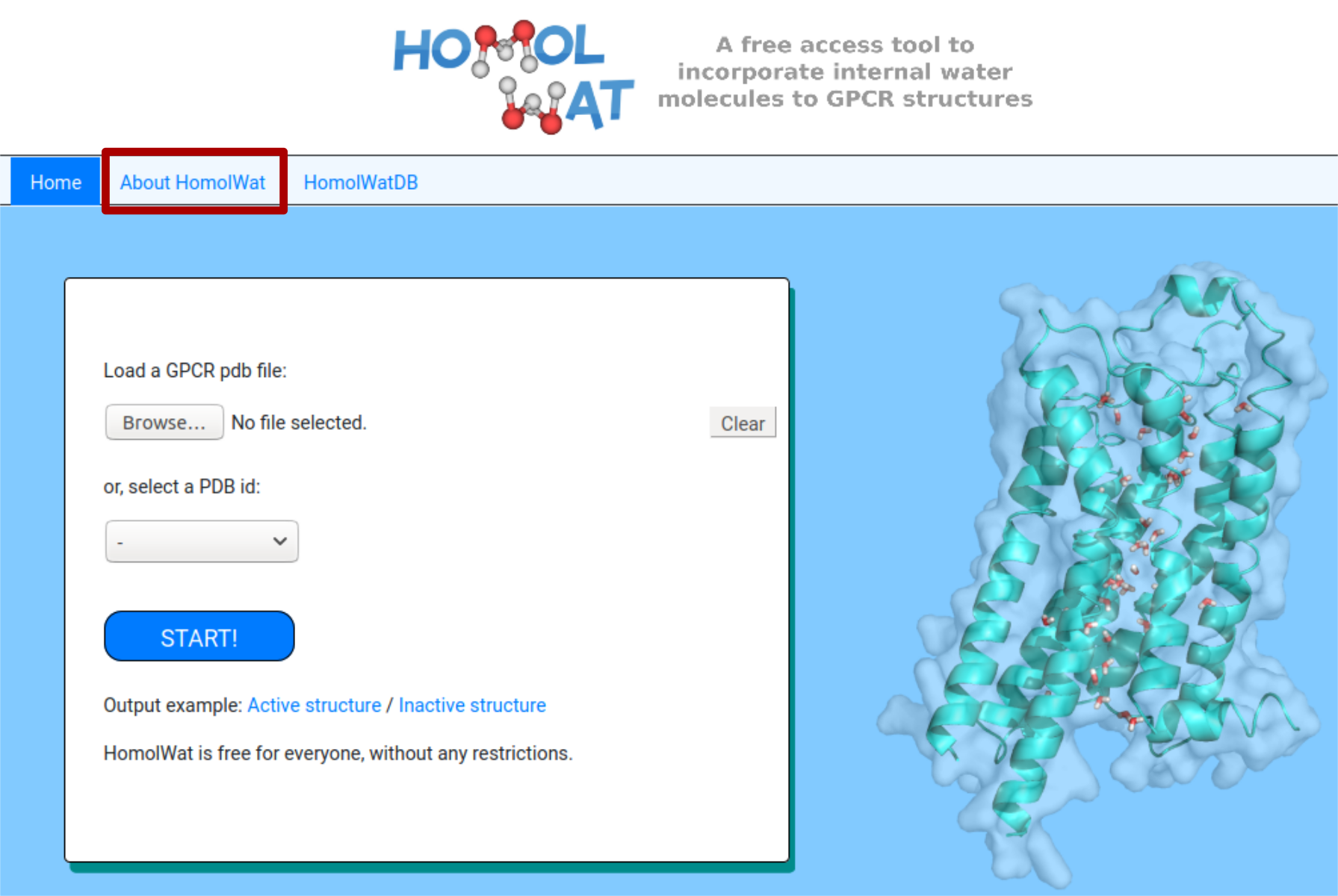
The first step is to provide the coordinates for the 3D structure of a GPCR (model or experimentally determined) in PDB format [1] or to select a PDB ID (and a chain) of a GPCR structure from the list [2]. The structures are taken from HomolwatDB (see below) and lack auxiliary proteins used to assist in structure stabilization were also removed or any non-protein molecules other than water, or GPCR orthosteric/allosteric ligands.HomolWat recognise ligands and other molecules from the model by the start of each row in the pdb file. Users must check that each row of the ligand starts with 'HETATM', otherwise it would be discarded.
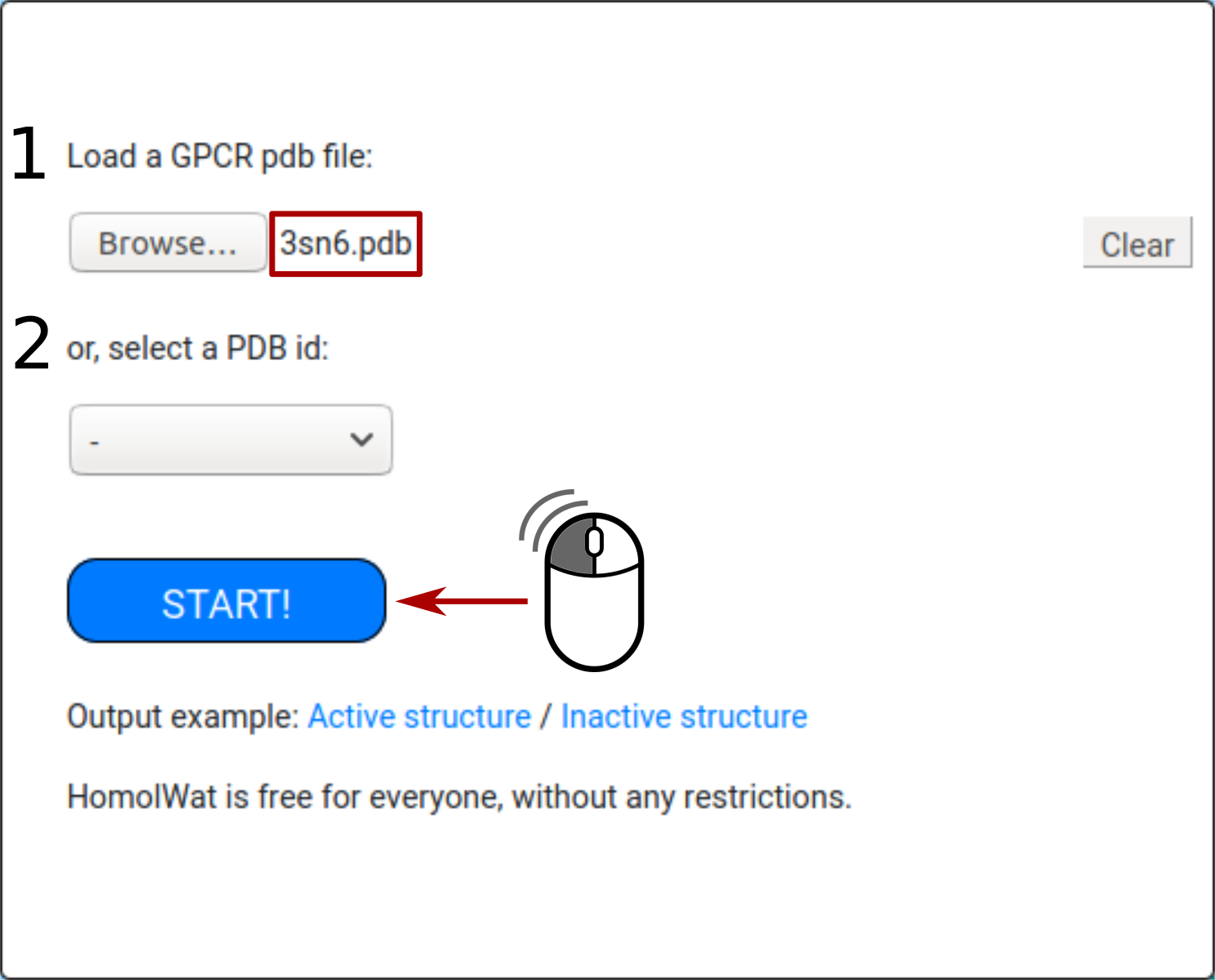
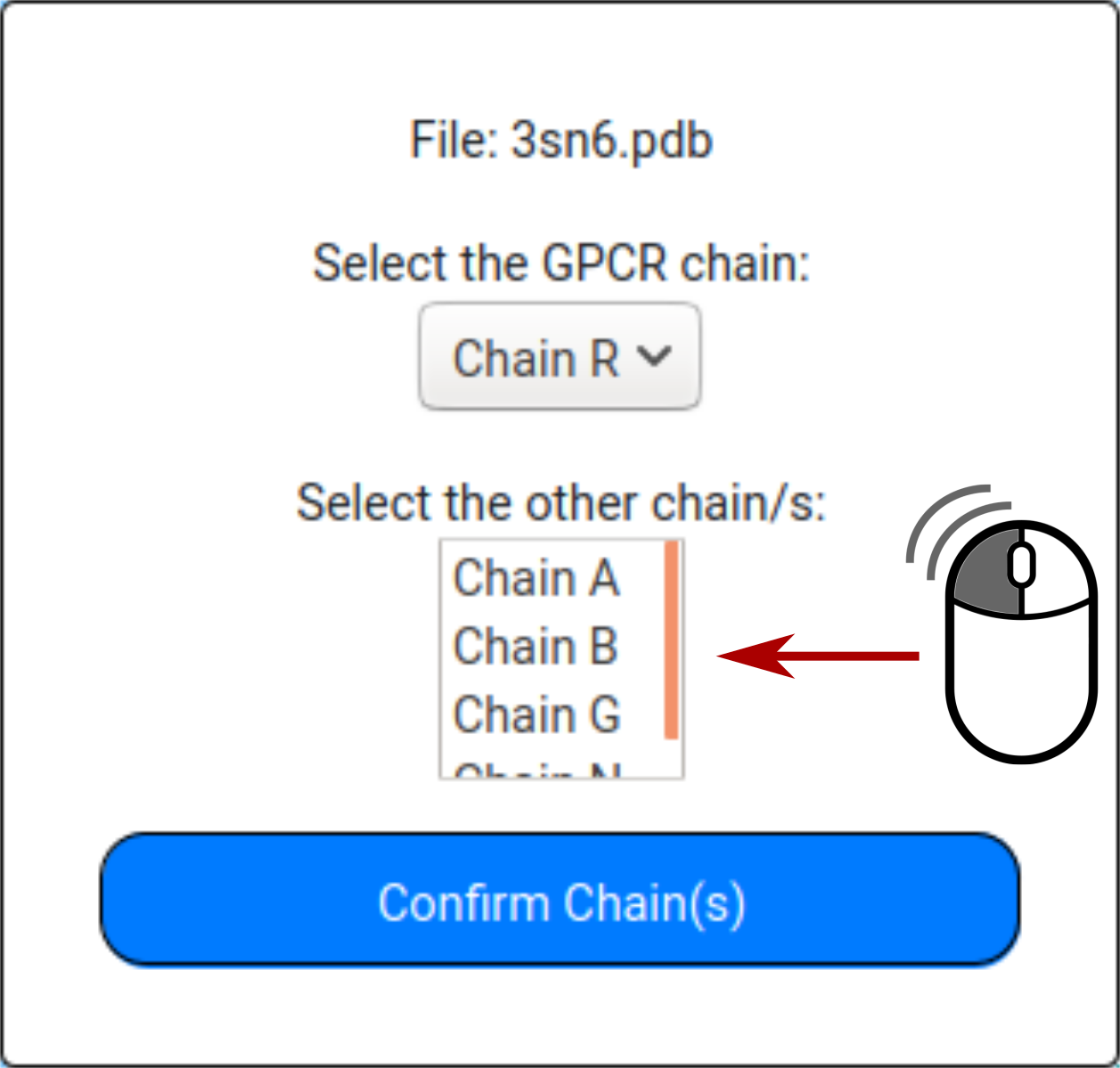
If more than one chain is available, the user can select the GPCR chain to be used to incorporate water molecules and other chains to be kept for water placement.
A table displays the blastp+ score, sequence identity, coverage and e-value of all proteins with internal water molecules that may be used for water placement.
The user can choose specific features such as:
-
1) Select the state (inactive or active) of the structure provided:
-
2) Incorporate or not an internal sodium ion (near residue D2.50). This is recommended for most inactive structures.
-
3) Running the protocol along with program Dowser+ (1). This option requires a few more minutes of computation. This option incorporates non-coincident water molecules predicted by Dowser+.
-
4) Minimum blastp+ score. By default HomolWat uses all high-resolution structures with water molecules available.
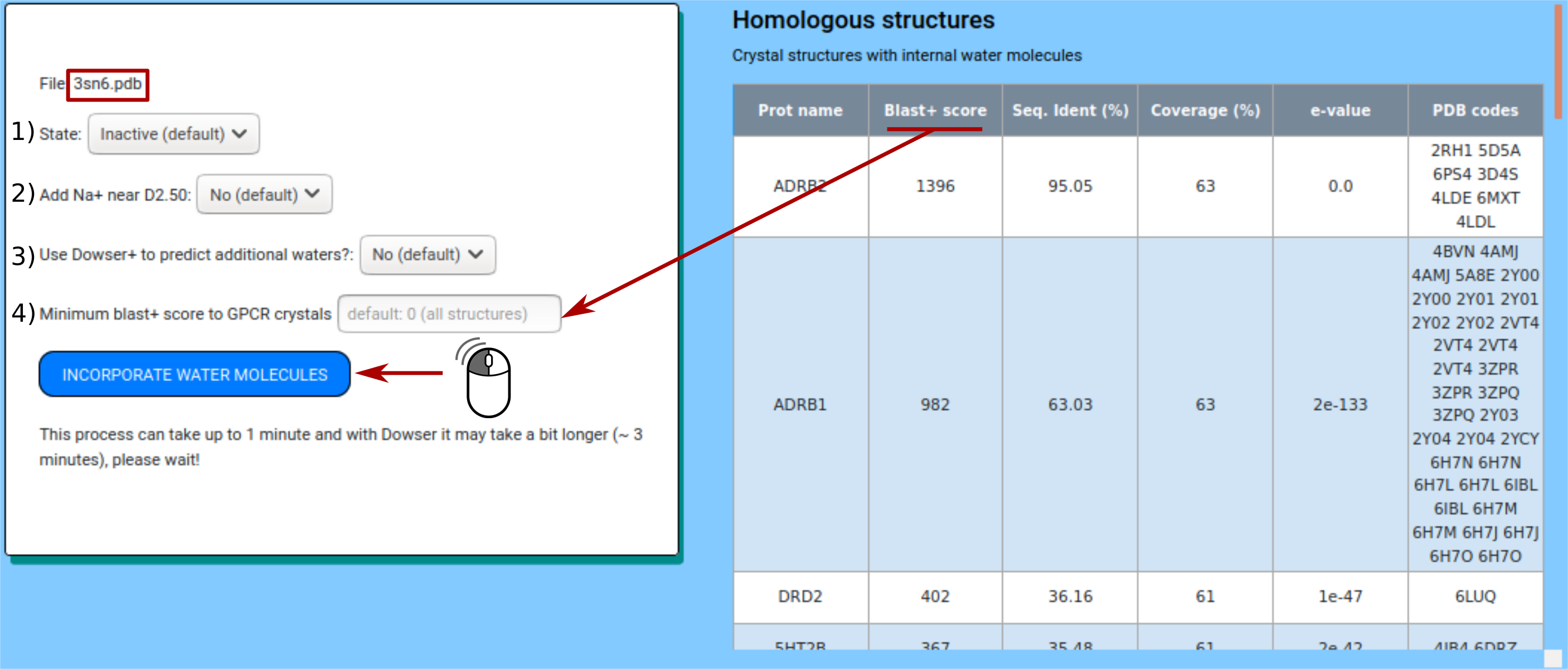
Clicking on ‘Incorporate water molecules’ button starts the protocol. The results will be presented back in about a minute. The final model can be visualized interactively with an NGL embedded viewer (2).
The final model can be downloaded in PDB format or in a PyMOL session that shows the origin the water molecules introduced. The output pdb file also includes the RMSD value for each water molecules according to the local structural alignment used to refine the water position. In addition, the user may download a csv file that contains for, each water molecule, the PDB id of the source structure and the RMSD of the global and local alignments.
HomolWatDB
HomolWatDB is the database of GPCR structures with internal water molecules that HomolWat relies on. This database can be browsed and downloaded. The structures can be visualized interactively within the HomolWat page using NGL (2).
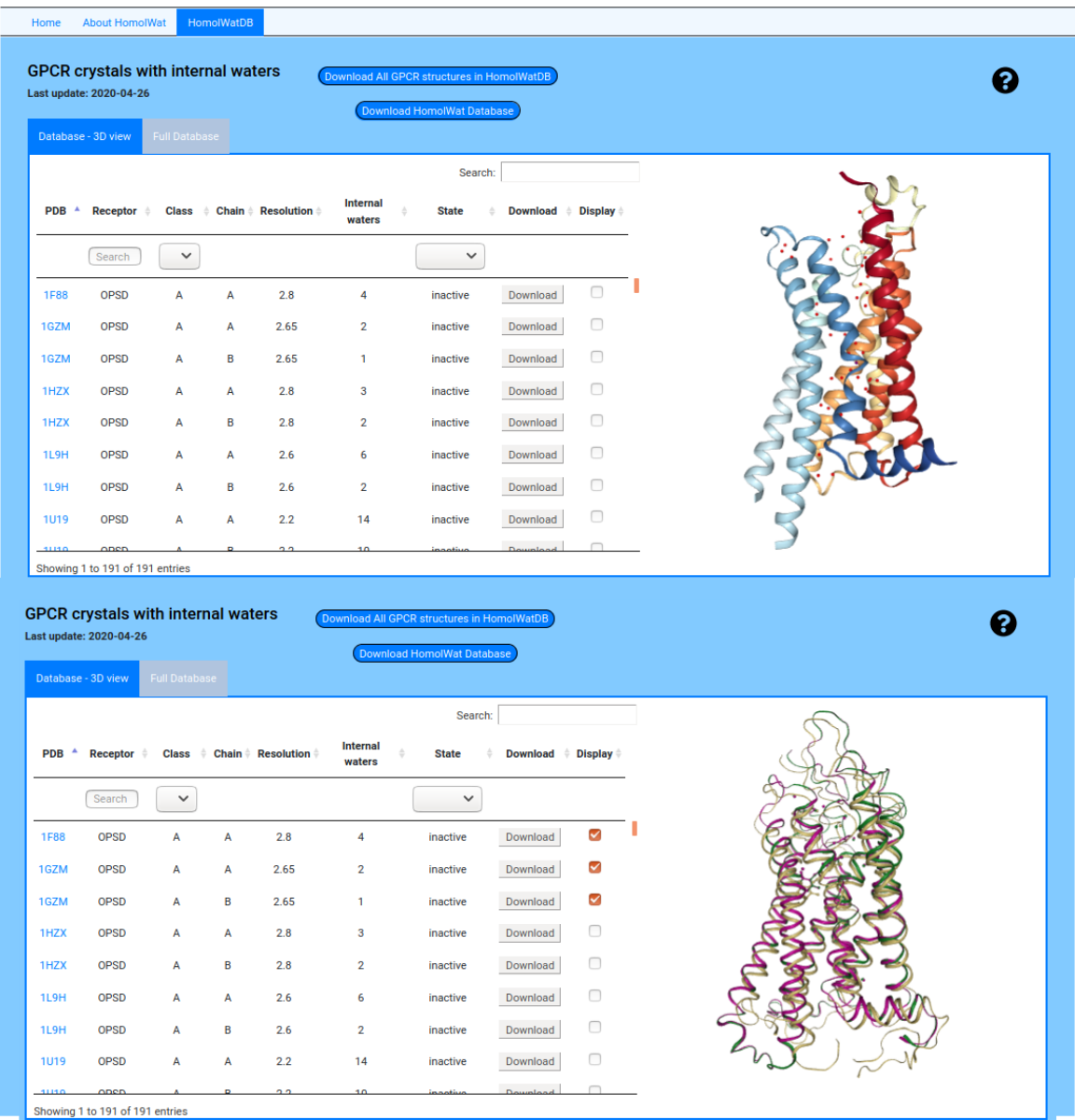
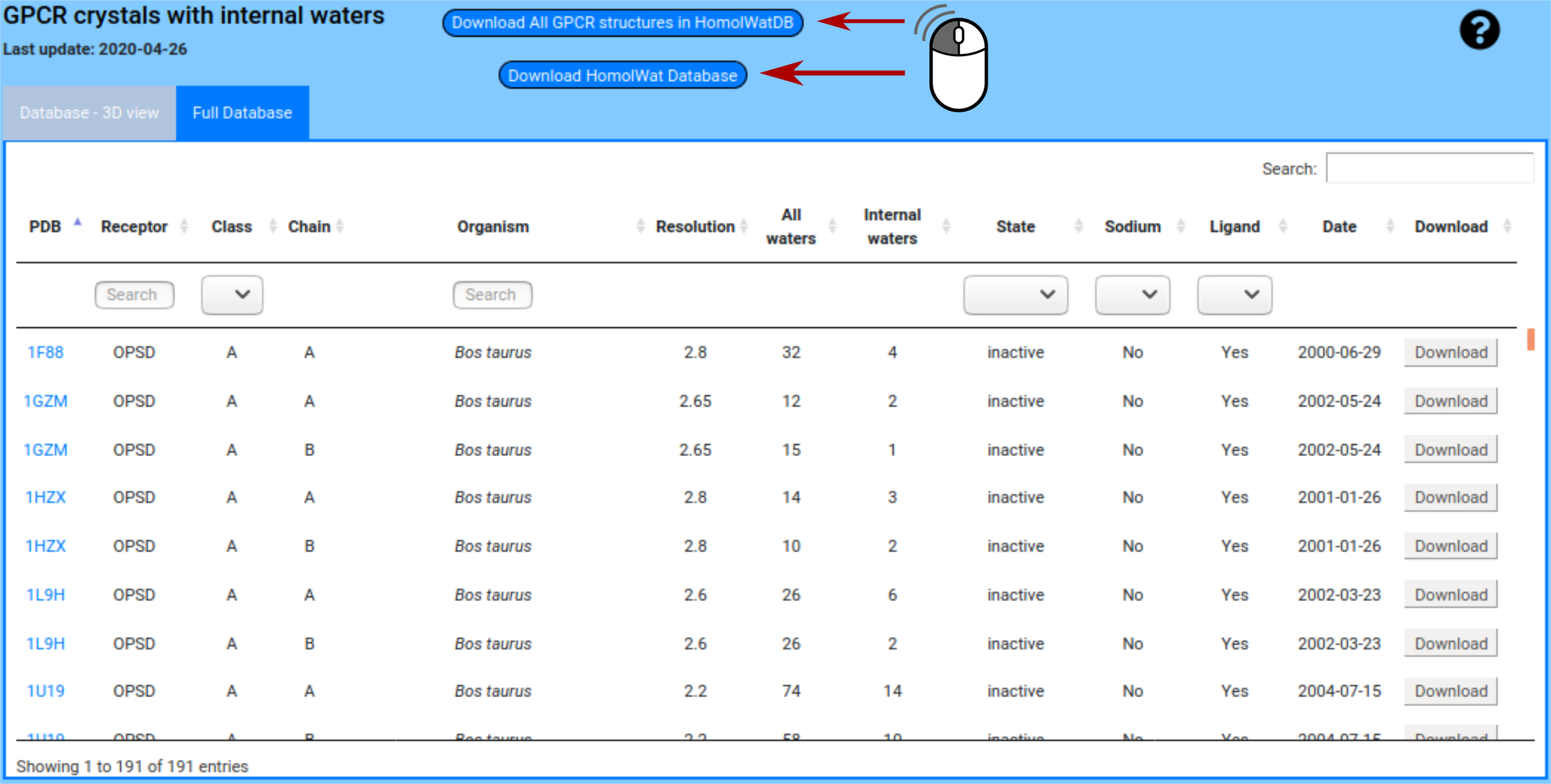
The full database table displays all the information of each structure. And also you can download the whole database in csv format or download all the structures in pdb format present in HomolWatDB.
Browser compatibility
| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
|---|---|---|---|---|---|
| Linux |
Ubuntu 18.04.3 |
78.0.3904.108 | 71.0 | n/a | n/a |
| MacOs | HighSierra 10.13.6 | 79.0.3945.88 | 71.0 | n/a | 13.0 |
| Windows | 10 | 78.0.3904.108 | 71.0 | 44.18362.449.0 | n/a |
Contact
Prepared by Eduardo Mayol, in collaboration with Adrián García, Mireia Olivella and Arnau Cordomí at
UAB and Peter Hildebrand and Ramon Guixa at Charite Berlin.
Admin contact e-mail: e.mayoles@gmail.com
References
(1) Morozenko, A., Leontyev, I. V., & Stuchebrukhov, A. A. (2014). Dipole moment and binding energy of water in proteins from crystallographic analysis. Journal of chemical theory and computation, 10(10), 4618-4623.
(2) Rose, A. S., & Hildebrand, P. W. (2015). NGL Viewer: a web application for molecular visualization. Nucleic acids research, 43(W1), W576-W579.
(3) Mezei, M. A new method for mapping macromolecular topography. J Mol Graph Model 2003, 21(5),
(4) Yuan, Z., Bailey, T. L., & Teasdale, R. D. (2005). Prediction of protein B‐factor profiles. Proteins:Structure, Function, and Bioinformatics, 58(4), 905-912.